



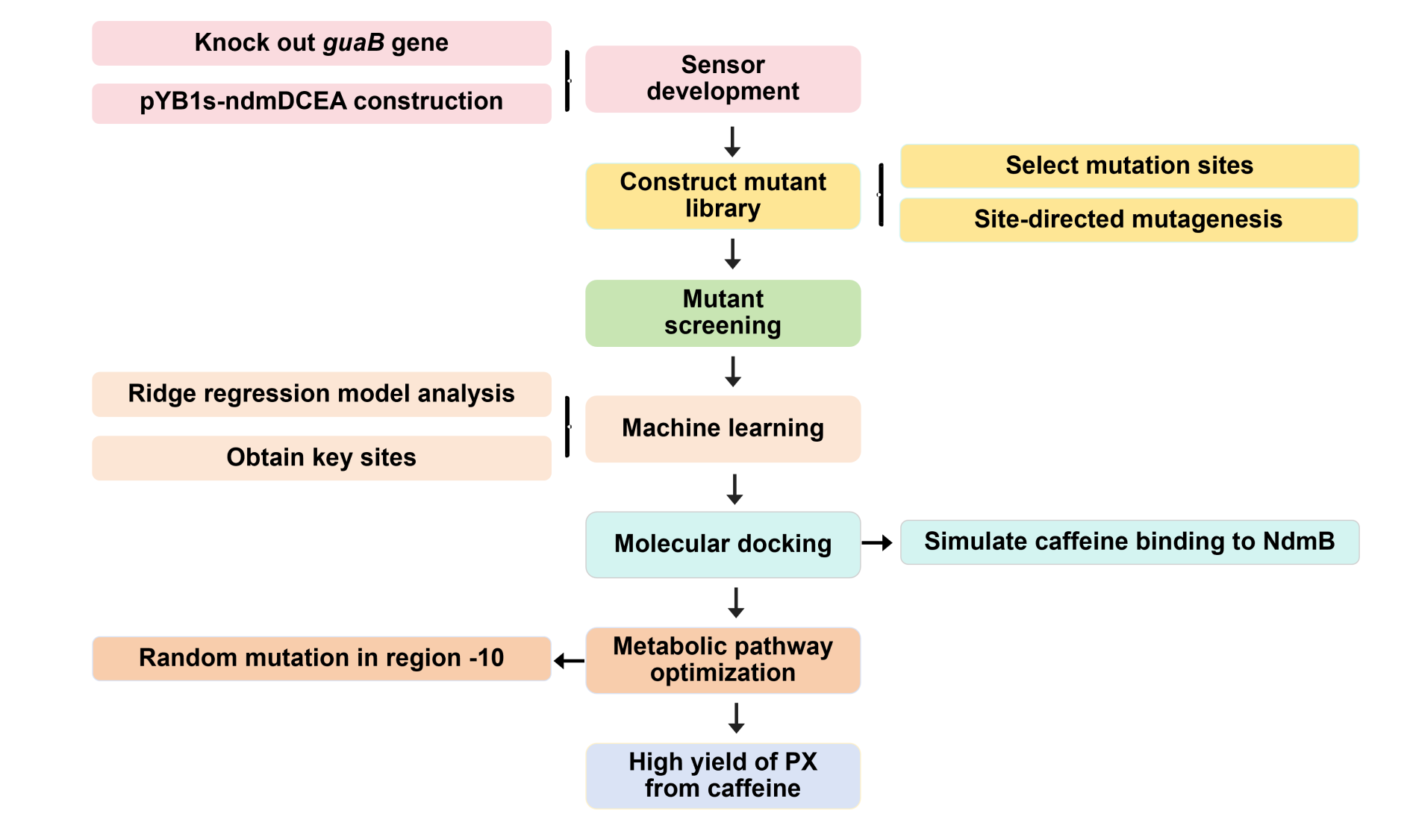
We plan to develop a whole-cell biosensor by knocking out the guaB gene in E. coli, which will block the synthesis pathways of DNA and RNA, preventing bacterial growth in the absence of exogenous xanthine. Additionally, the ndmDCEA gene cluster was introduced, where ndmA catalyzes N1-demethylation and ndmC catalyzes N7-demethylation, which can promote the conversion of PX into xanthine. Bacterial survival depends on efficient PX production by NdmB-catalyzed N3-demethylation of caffeine.
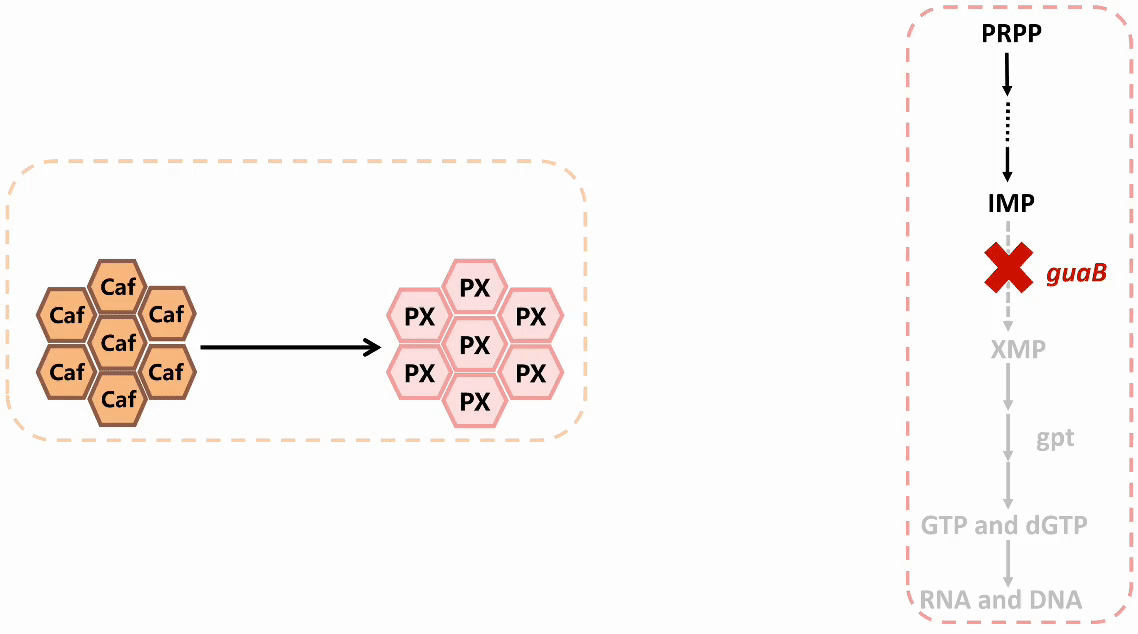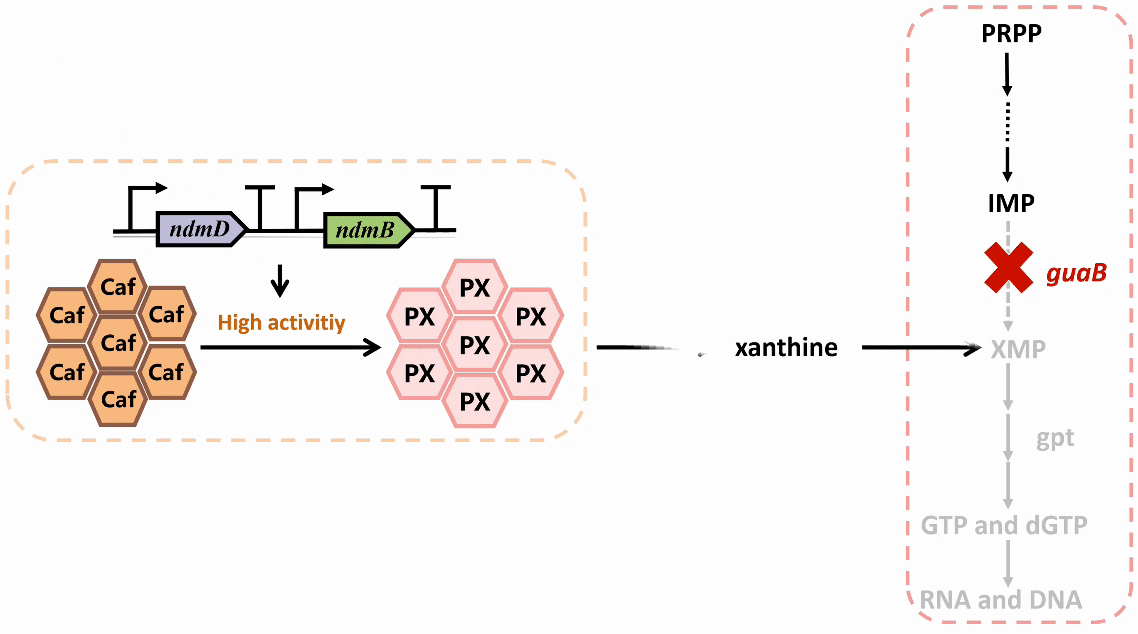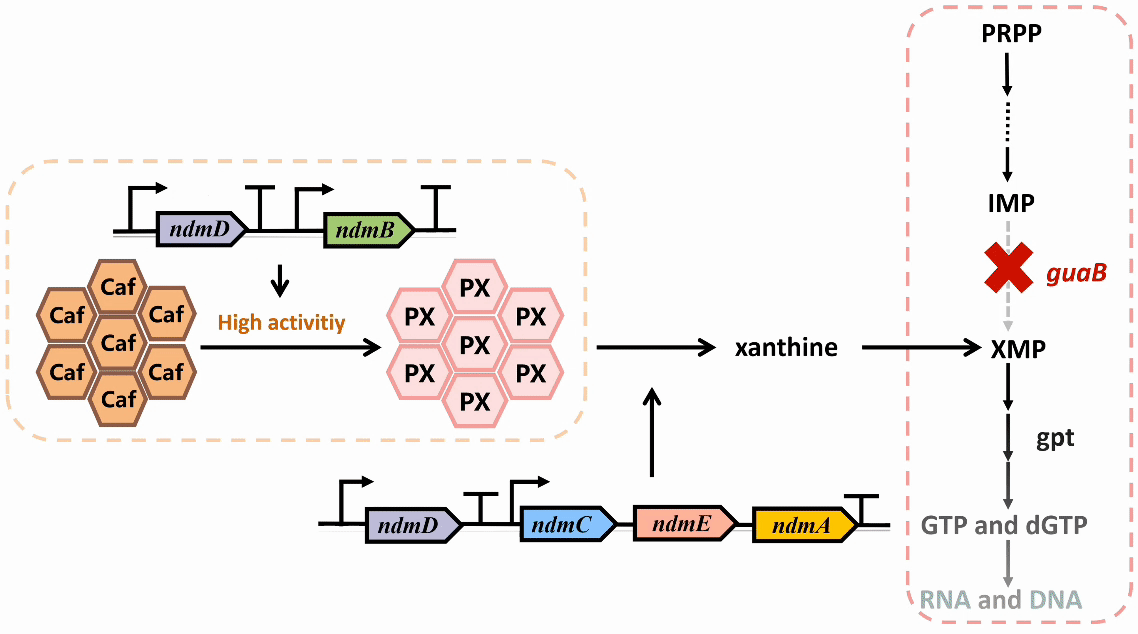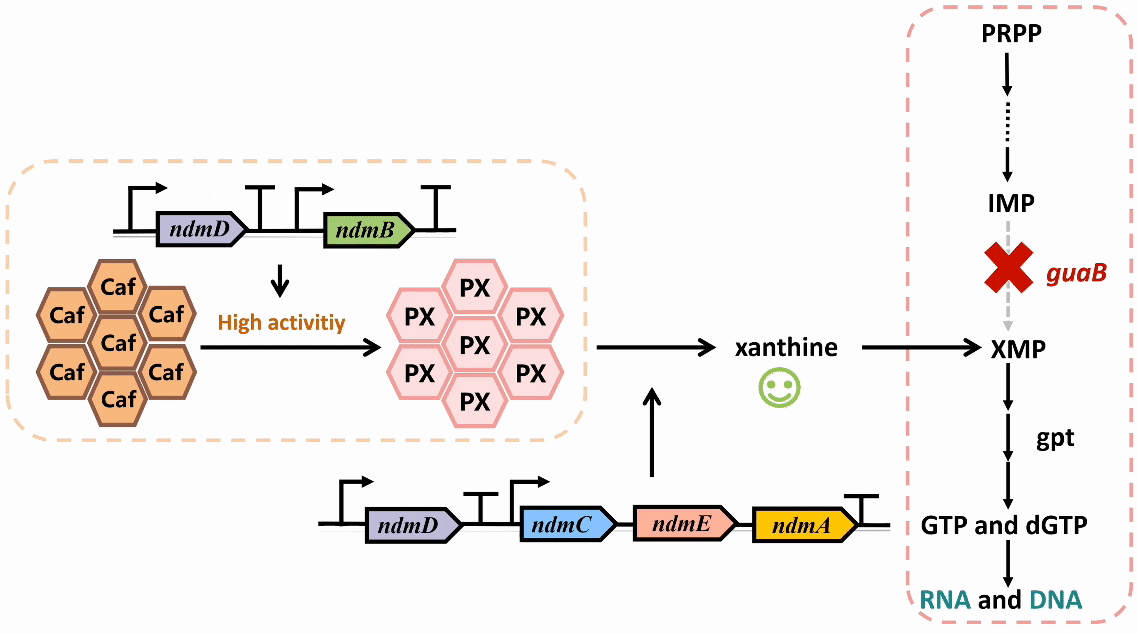
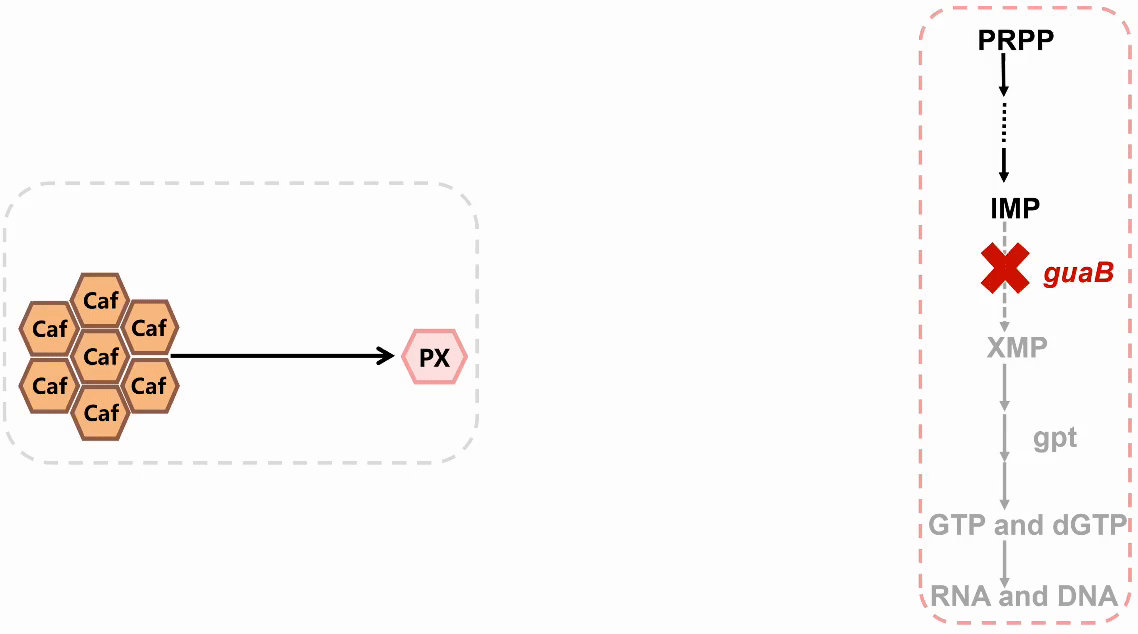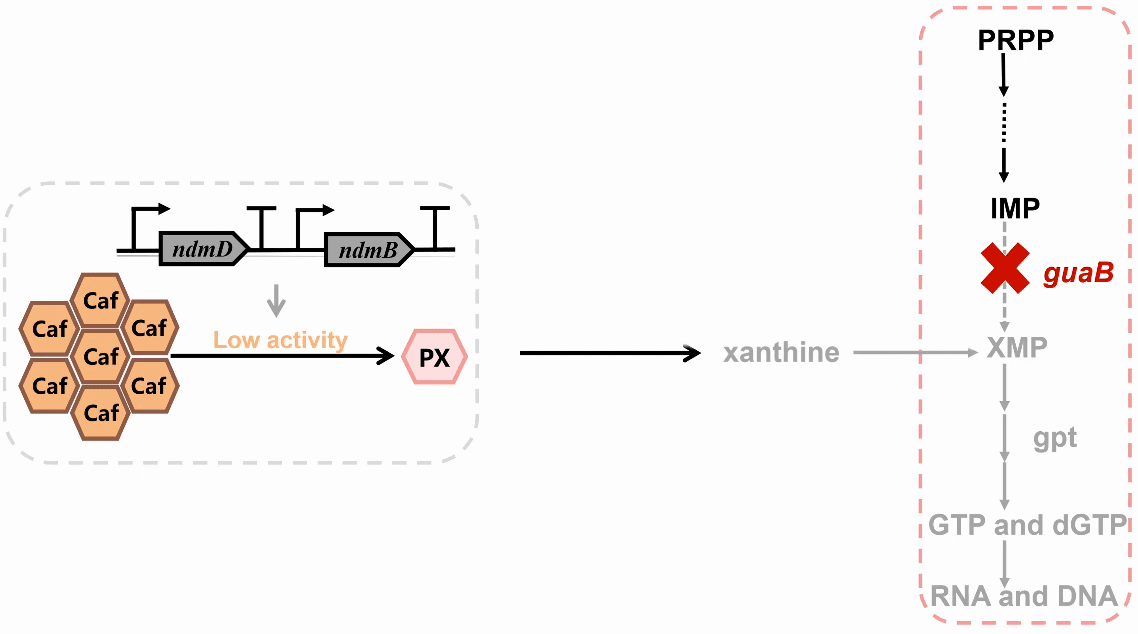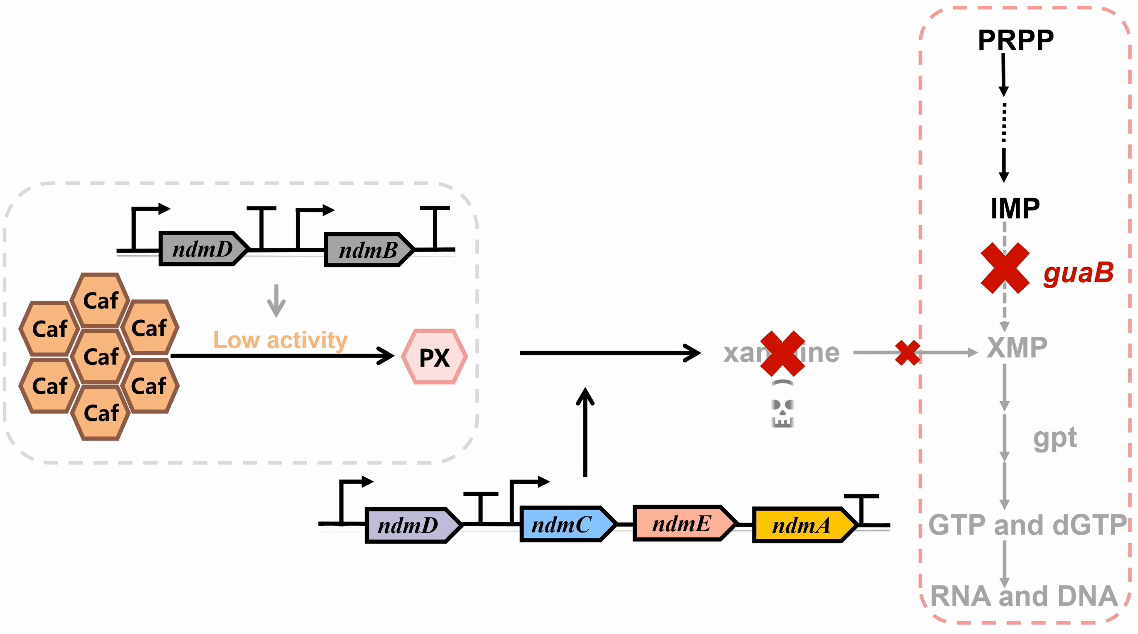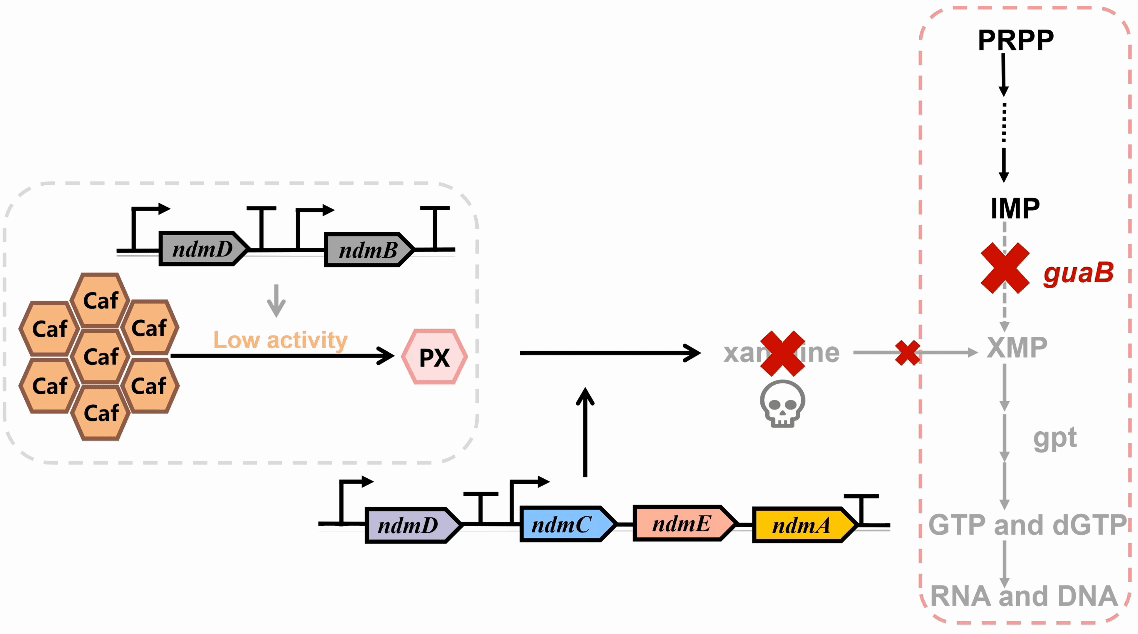
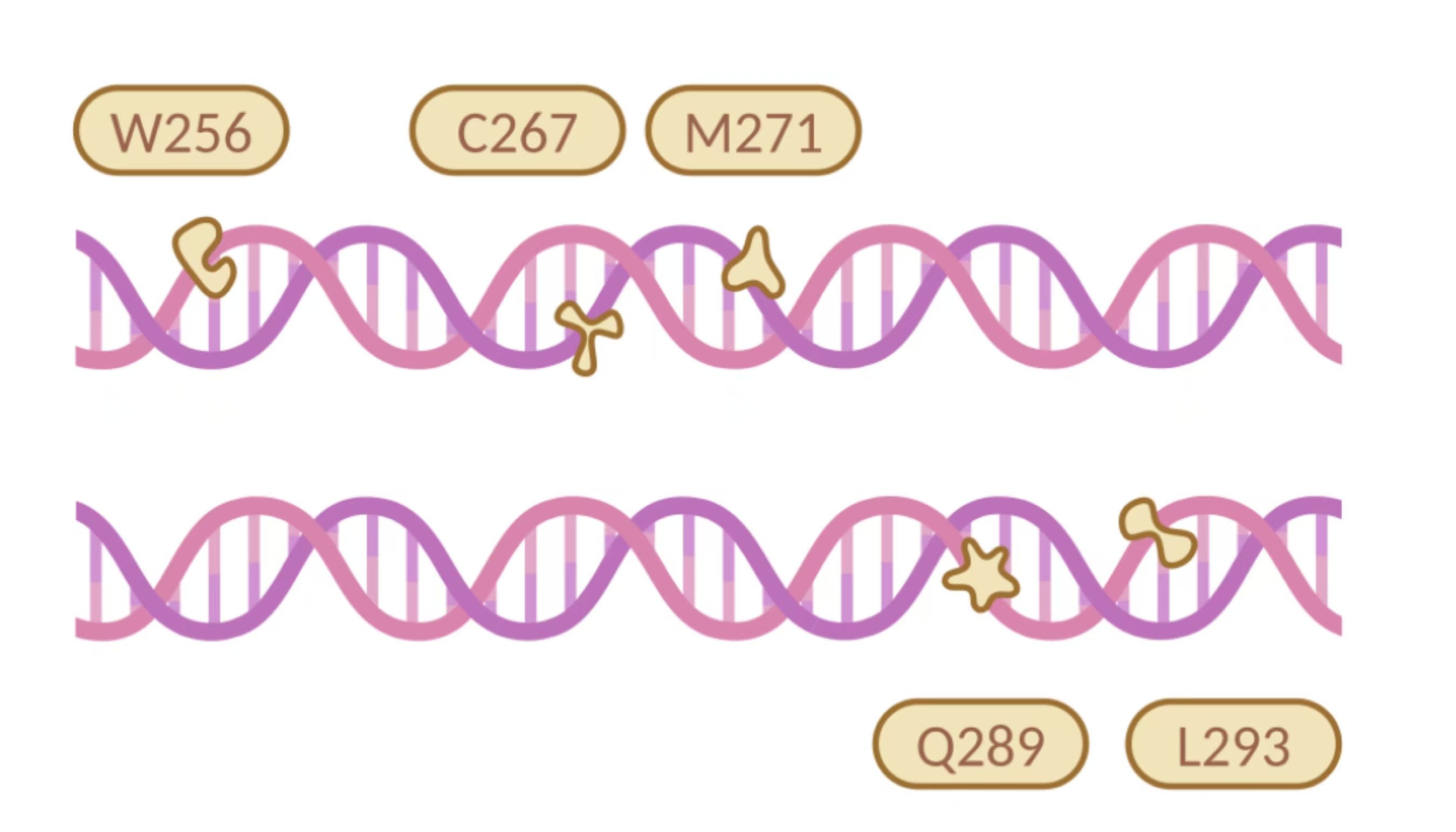
In the selection of mutation sites, previous studies have analyzed the substrate binding interactions within the SRPBCC domain of the NdmA enzyme for both caffeine and theobromine, identifying key residues (Q289 and L293) as critical for substrate specificity1. Homology modeling of NdmA and NdmB also revealed non-conserved residues within the active site pocket (W256, C267, and M271), which could likely contribute to substrate binding orientation and specificity2. Based on this prediction, we performed combinatorial mutations at two sets of residues and employed the biosensor plasmid to screen for clones based on their viability.
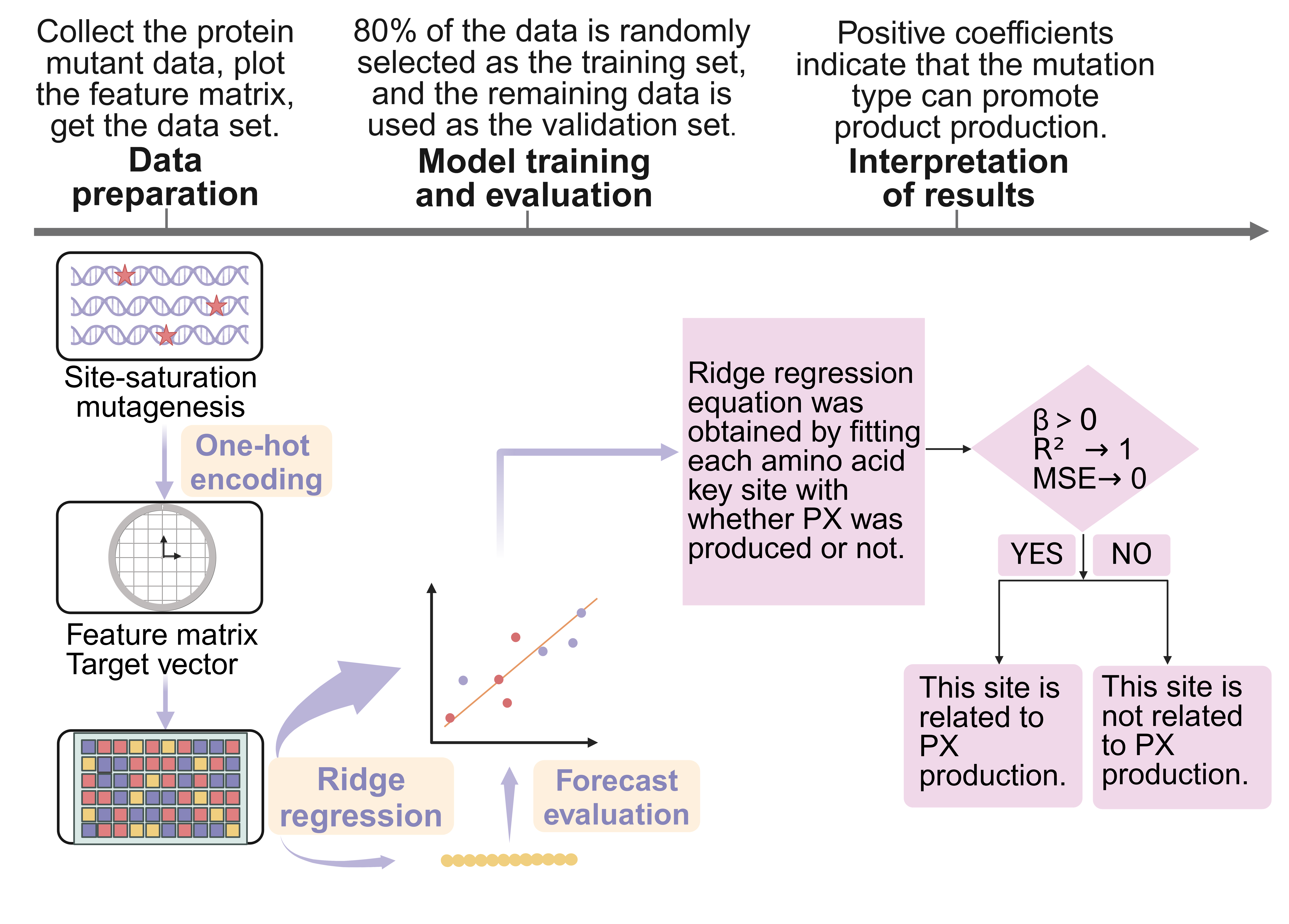
To enhance the efficiency of protein modification and explore the relationship between protein mutations and the generation of multiple products, thereby identifying mutation sites that favor the production of pseudoxanthine, we plan to utilize a Ridge regression model for machine learning.
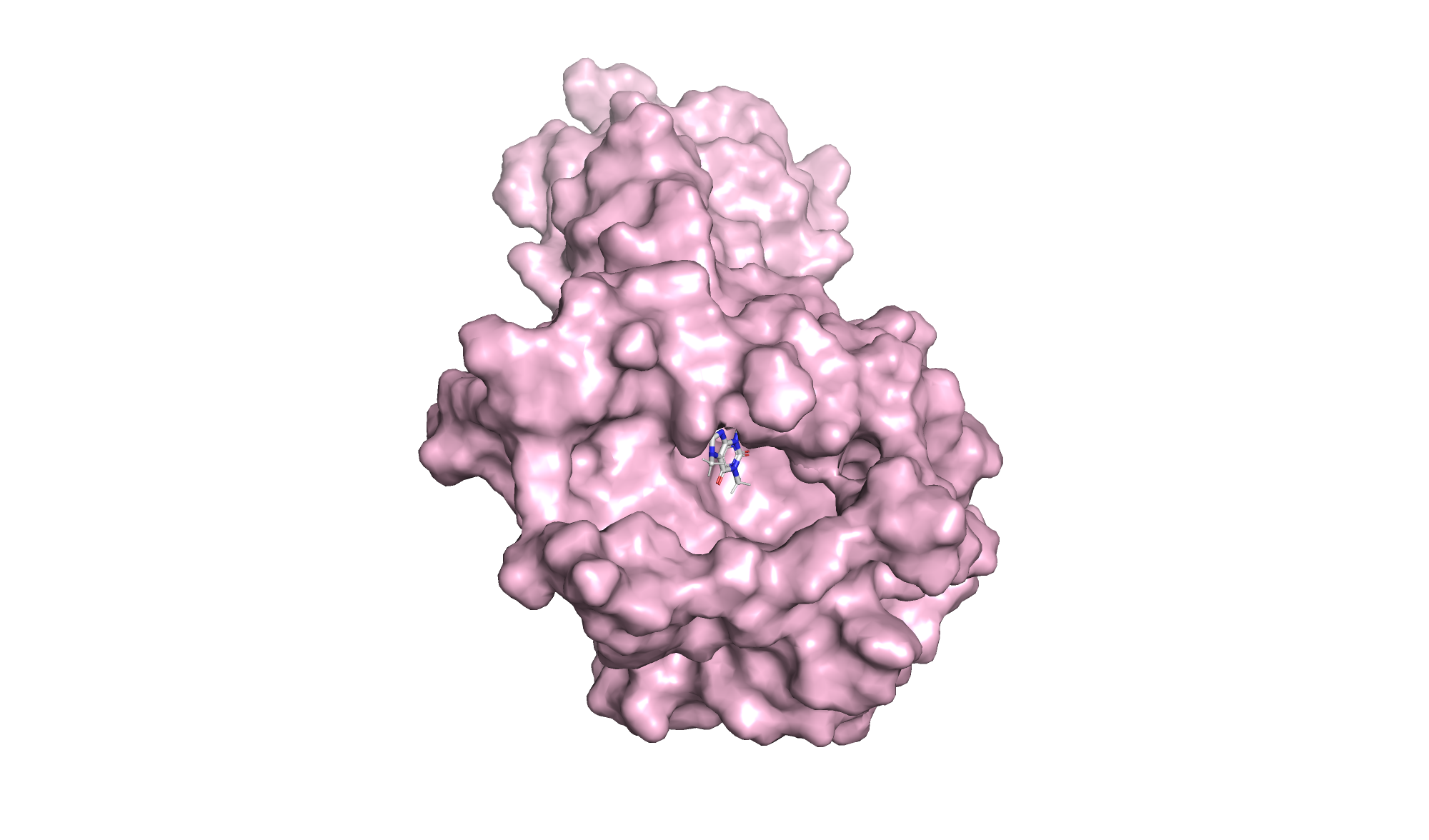
To accurately elucidate the authentic state of the target protein under whole-cell catalytic conditions, the NdmB structure retrieved from the Protein Data Bank (PDB) will be subjected to optimization via molecular dynamics simulations. Upon generating protein models for both the wild-type NdmB and its variants, AutoDock Vina will be employed to investigate the docking interactions between caffeine and NdmB, with the resulting docking conformations subsequently visualized using PyMOL. Additionally, a comparative analysis of the structural dynamics and interactions of both the wild-type NdmB and its mutants in the presence of caffeine will be performed utilizing GROMACS.
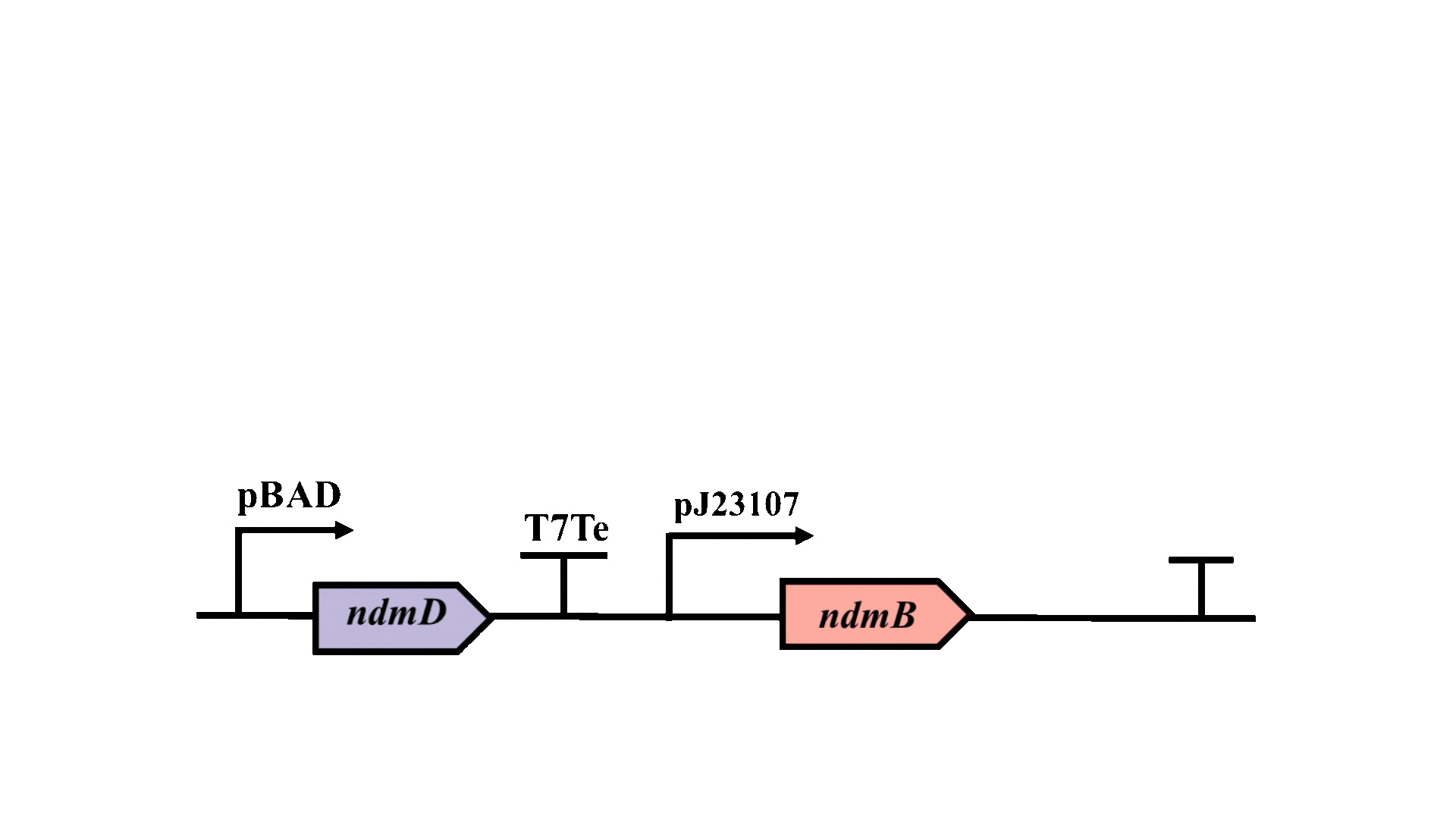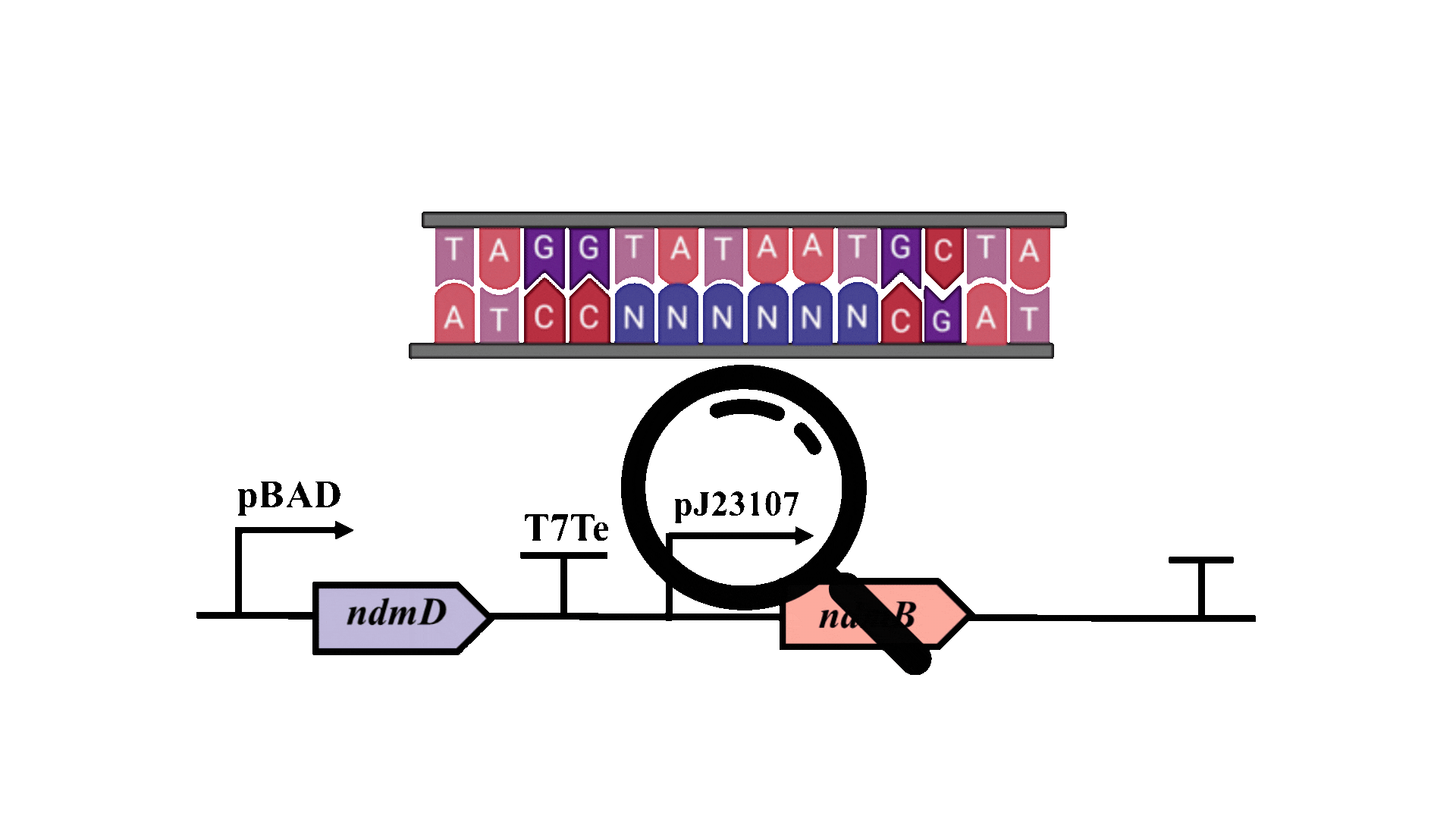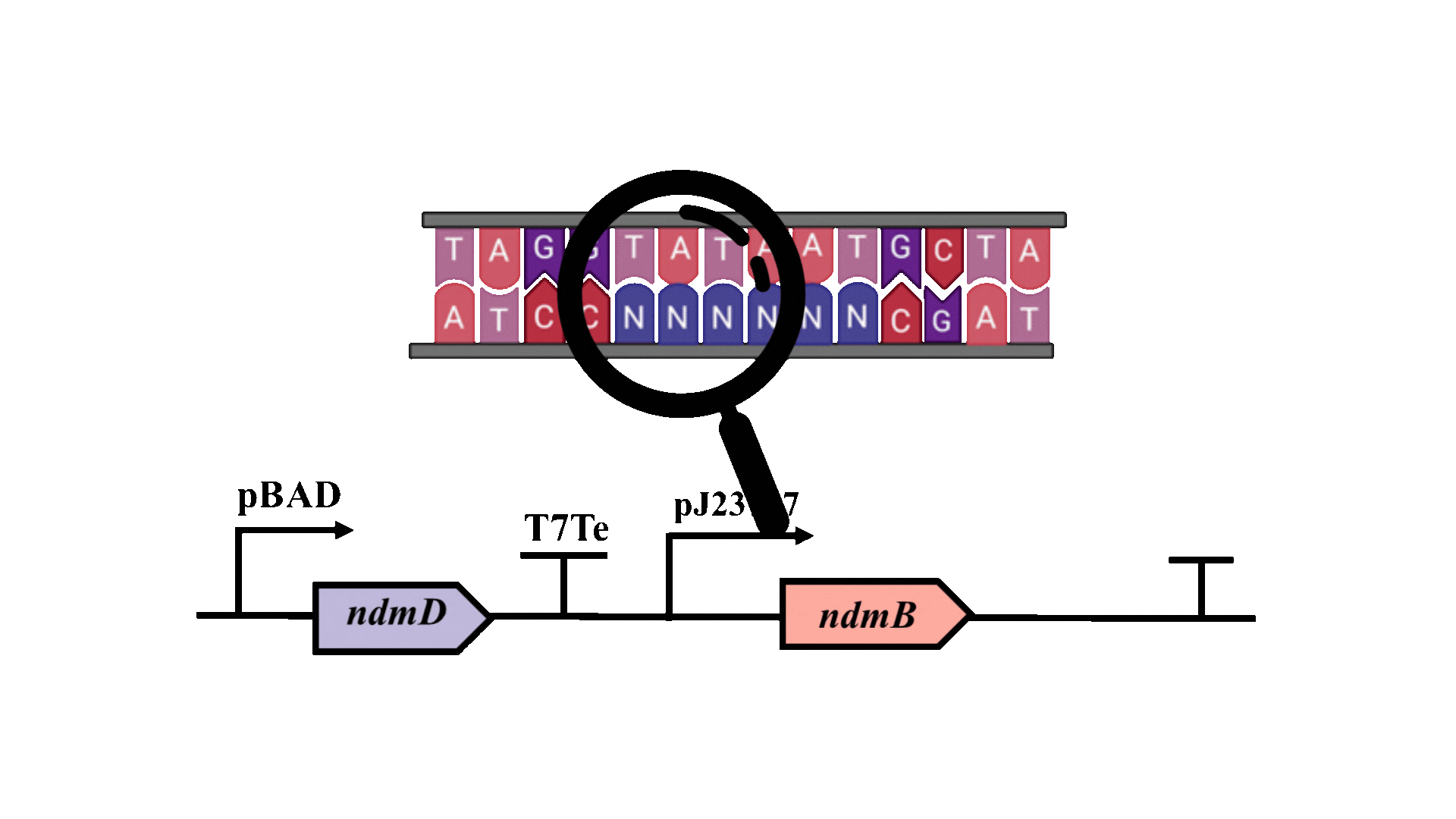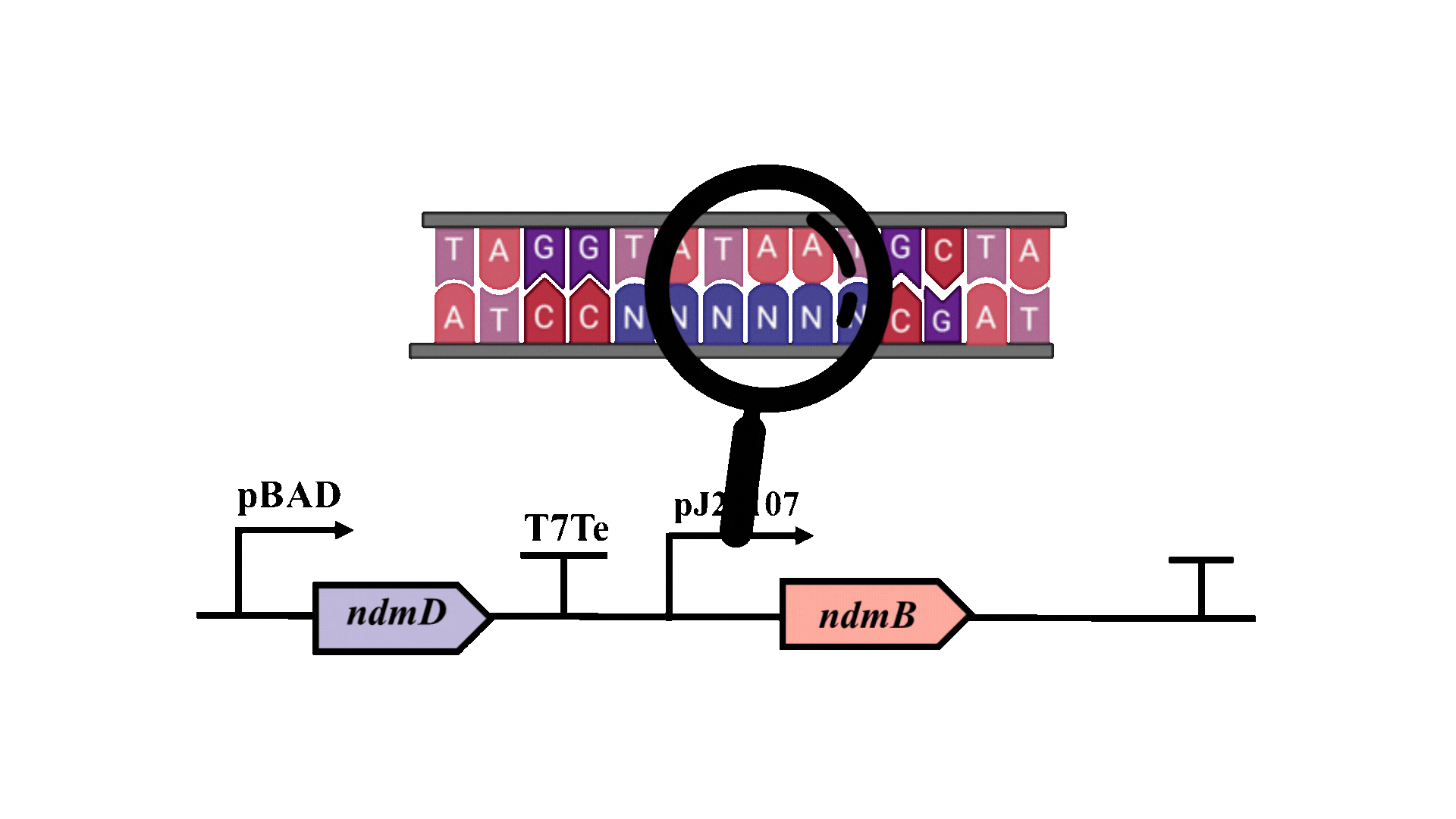
The possibility of further optimizing the yield is being considered, with particular emphasis on enhancing the metabolic pathway. Promoter optimization is crucial for improving the metabolic pathway, as the -10 region is essential for the binding of RNA polymerase to DNA and the initiation of transcription3. Consequently, degenerate primers will be employed to introduce random mutations at six bases within the -10 region of the pJ23107 promoter, and a whole-cell biosensor will be utilized for screening and analysis.
1. Summers, R. M., Louie, T. M., Yu, C., Gakhar, L., Louie, K. C., & Subramanian, M, Novel, highly specific N -Demethylases enable bacteria to live on caffeine and related purine alkaloids. Journal of Bacteriology, 194(8), 2041–2049, (2012).
DOI: https://doi.org/10.1128/jb.06637-11
2. Kim, J. H., Kim, B. H., Brooks, S., Kang, S. Y., Summers, R. M., & Song, H. K, Structural and Mechanistic Insights into Caffeine Degradation by the Bacterial N-Demethylase Complex. Journal of Molecular Biology, 431(19), 3647–3661, (2019).
DOI: https://doi.org/10.1016/j.jmb.2019.08.004
3. Zhou, C., Ye, B., Cheng, S., Zhao, L., Liu, Y., Jiang, J., & Yan, X, Promoter engineering enables overproduction of foreign proteins from a single copy expression cassette in Bacillus subtilis. Microbial Cell Factories, 18(1), 111–121, (2019).
DOI: https://doi.org/10.1186/s12934-019-1159-0